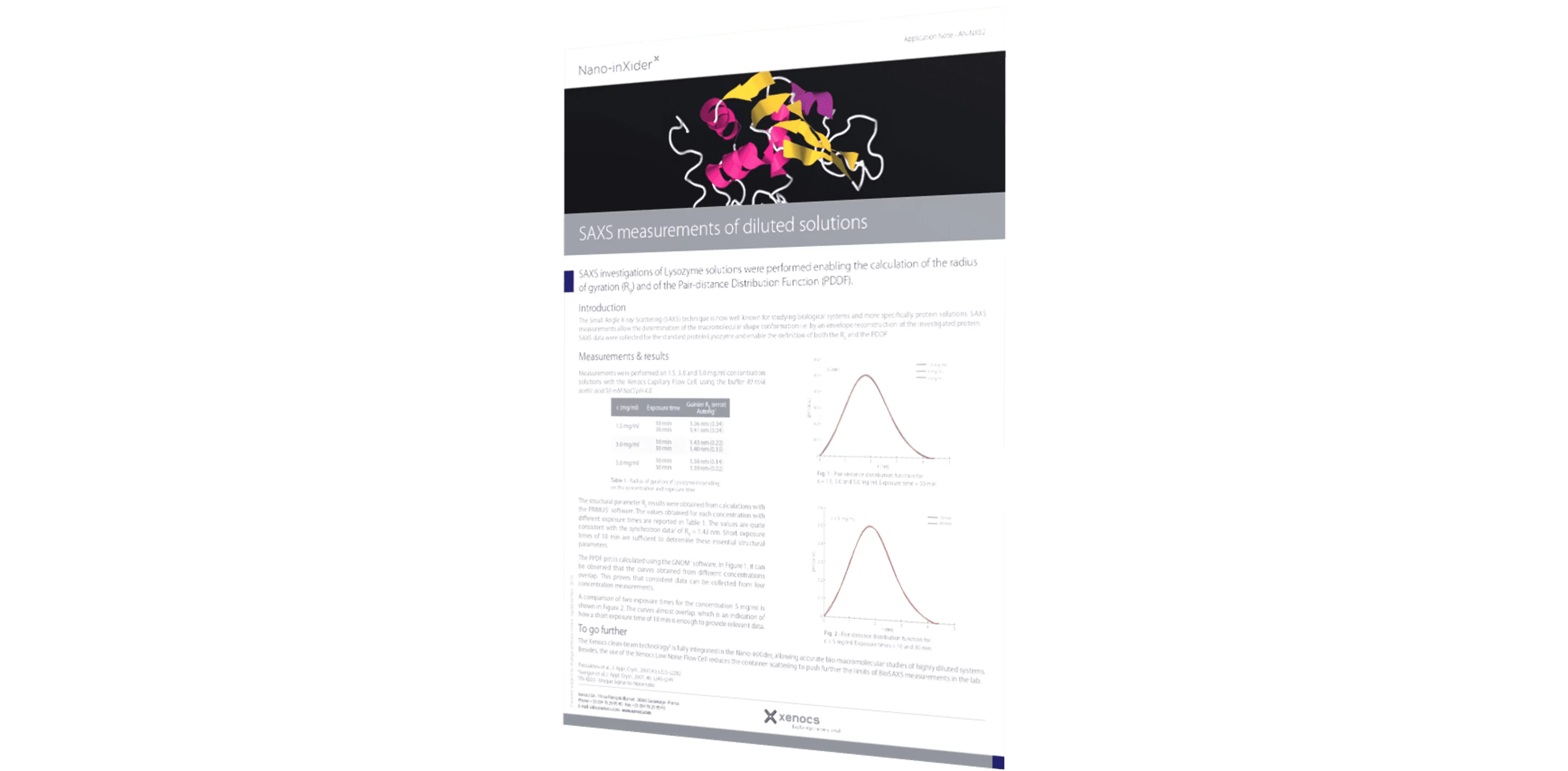
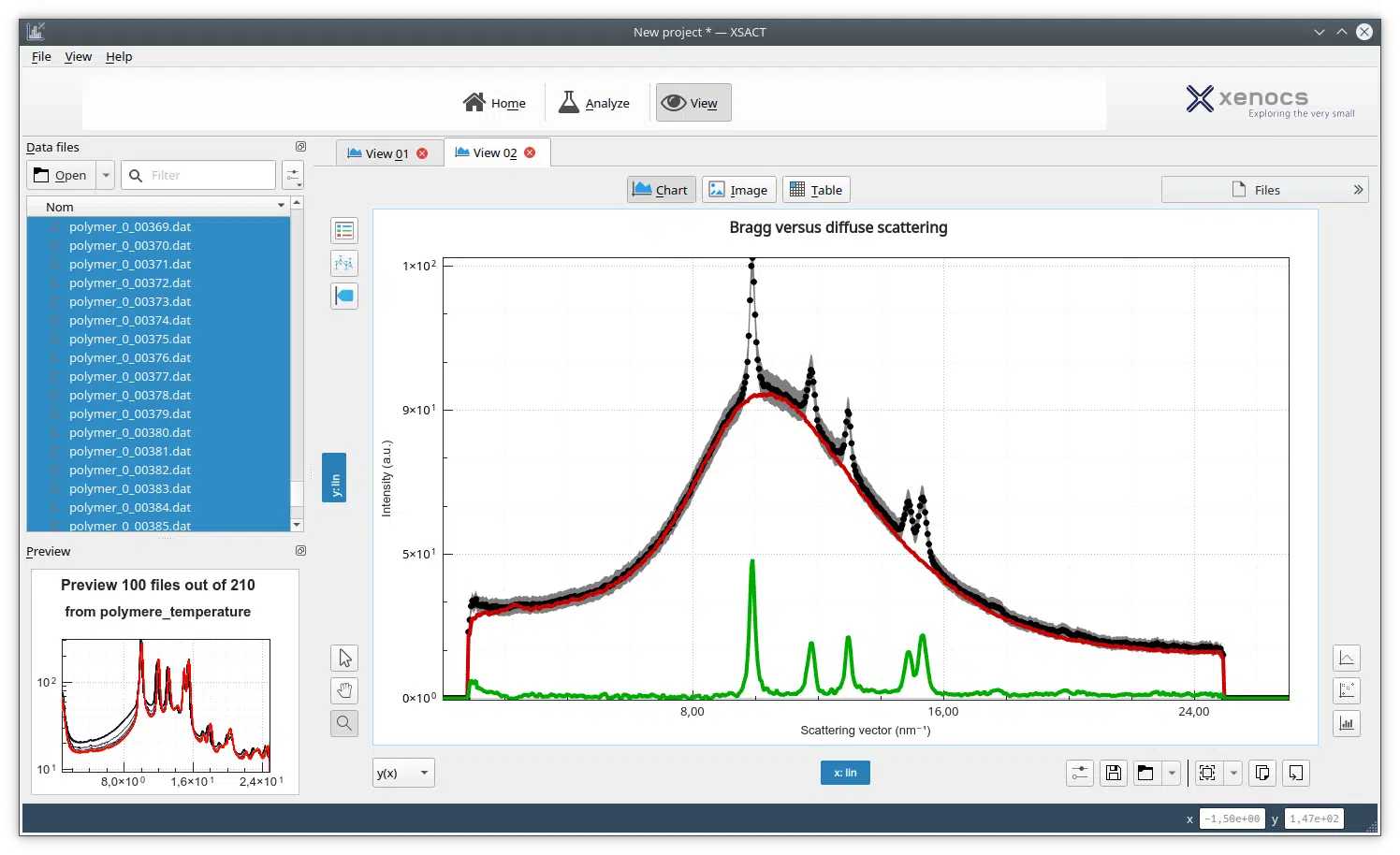
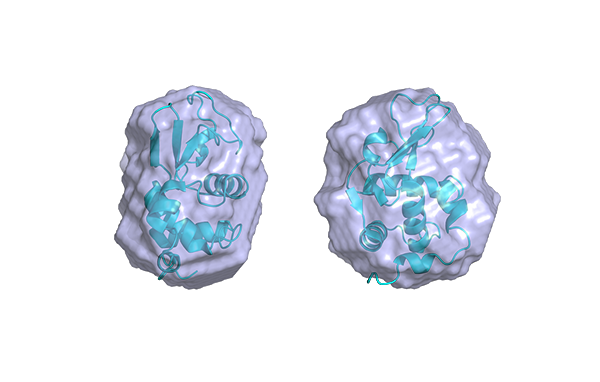
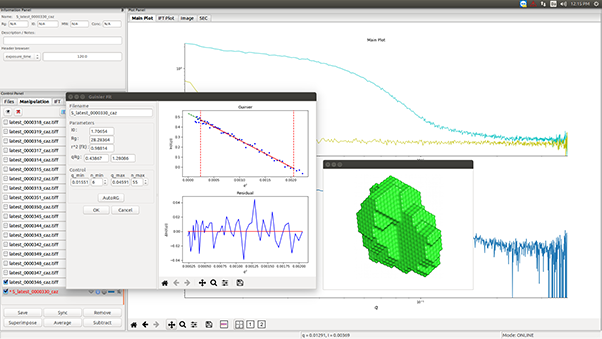
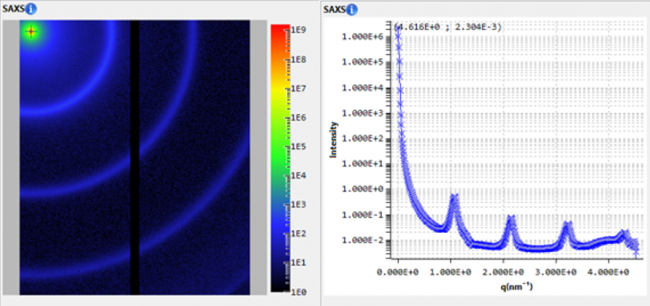
SAXS investigations of Lysozyme solutions were performed enabling the calculation of the radius of gyration (Rg) and of the Pair-distance Distribution Function (PDDF).
Introduction
Small Angle X-ray Scattering (SAXS) has become an established technique for studying biological systems and more specifically protein solutions. SAXS is able to characterize both ordered and disordered proteins in solution. It provides information about the shapes and sizes of proteins and complexes in a broad range of molecular sizes and under various experimental conditions ranging from extreme (e.g. low temperatures or high pressure) to nearly native.1
From the SAXS scattering pattern, there are several characteristic parameters of the investigated sample that can be obtained such as the radius of gyration, volume, the maximum dimension (Dmax), the folding state or the molecular weight. Rg provides a measure of the overall size of the macromolecule and it can be estimated using the Guinier approximation or from the pair-distance distribution function which is a histogram of distances between all possible pairs of atoms within a particle.1
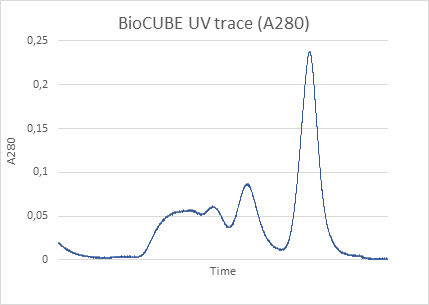
The scattering intensity in a SAXS experiment is proportional to the concentration of the solution: the higher the concentration the better the signal-to-noise ratio of the solvent-subtracted data. It can thus be challenging to perform macromolecular studies of highly diluted systems.
In this application note, we used small angle X-ray scattering data collected on the standard protein Lysozyme in different concentrations and using long and short exposure times. We show that it is possible to obtain accurate information on the PDDF and Rg values with a laboratory based SAXS instrument, consistent with synchrotron data2 in as little as 10 minutes exposure time and for concentrations as low as 1.5 mg/ml.
1Kikhney, A. G., & Svergun, D. I. (2015). A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS letters, 589(19), 2570-2577. doi: 10.1016/j.febslet.2015.08.027.
2 Mylonas, E., & Svergun, D. I. (2007). Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering. Journal of applied crystallography, 40(s1), s245-s249. doi: 10.1107/S002188980700252X
.