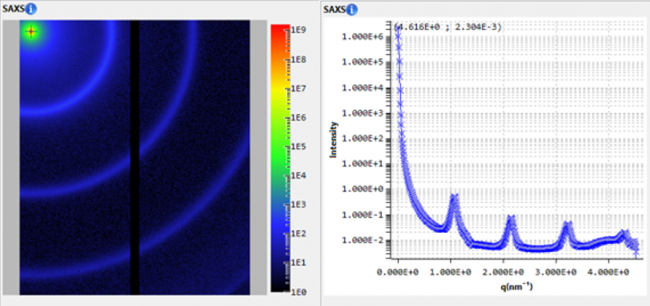
Investigating melittin self-assembly in solution using laboratory SAXS
Comprehensive knowledge of the structure and solution-state behavior of peptides is essential for understanding their biological functions. Various techniques, such as crystallography, electron microscopy (EM), nuclear magnetic resonance (NMR), and dynamic light scattering (DLS) provide valuable structural and biophysical data. However, these methods often face challenges when it comes to small peptides, particularly those that are flexible, glycosylated, difficult to crystallize, or intrinsically disordered [1-2]. Small -Angle X-ray Scattering (SAXS) offers a versatile alternative that overcomes many of these limitations. SAXS enables the analysis of small peptides, even in low-molecular-weight oligomeric forms in a near-native solution environment. Unlike NMR or crystallography, SAXS can handle a wide range of sample sizes and conformational states, providing detailed insights into size, shape, oligomerization, stability, and interparticle interactions.
While DLS is commonly used to measure the hydrodynamic size and assess aggregation, it offers less structural information compared to SAXS. SAXS provides not only size information but also detailed insights into the overall shape, folding and flexibility of the molecules [2], which is invaluable for understanding complex assembly states in solution. Additionally, SAXS boasts high-throughput capabilities, enabling rapid data acquisition and analysis that exceeds the sample preparation and data interpretation times required by crystallography or labeling processes in NMR [3]. Furthermore, SAXS is a powerful tool for validating and refining models obtained from other techniques, such as crystallography, NMR, and computational structure predictions [4].