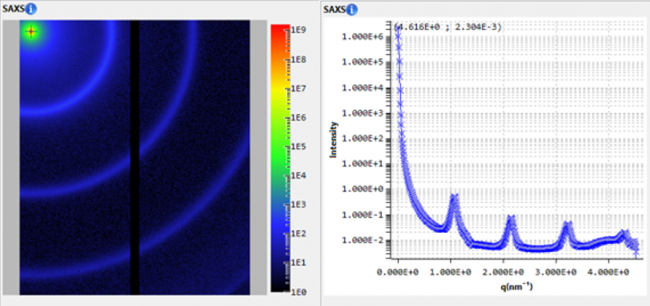
Laboratory SAXS was used to characterize monoclonal antibody in solution
Biotherapeutics, particularly monoclonal antibodies (mAbs), are well-established therapies recognized for their high specificity and adaptability in targeting various diseases, including cancers and autoimmune disorders [1, 2]. Detailed structural characterization is crucial for ensuring the stability, efficacy, and safety of mAbs throughout the drug development process [3-6]. Maintaining structural integrity under varying conditions—such as concentration, ionic strength, and formulation changes—is essential, as instability can lead to aggregation, degradation, or reduced binding affinity, thereby compromising therapeutic potential [5-7].
Optimizing antibody formulations and storage conditions requires a thorough understanding of their behavior in different physicochemical environments [8]. Factors such as concentration and ionic strength significantly influence conformational stability, intermolecular interactions, and aggregation tendencies [6, 8]. Addressing these challenges early in the development process is key to achieving long-term stability and therapeutic efficacy.
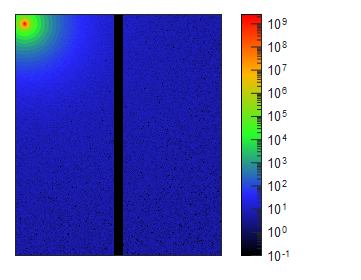
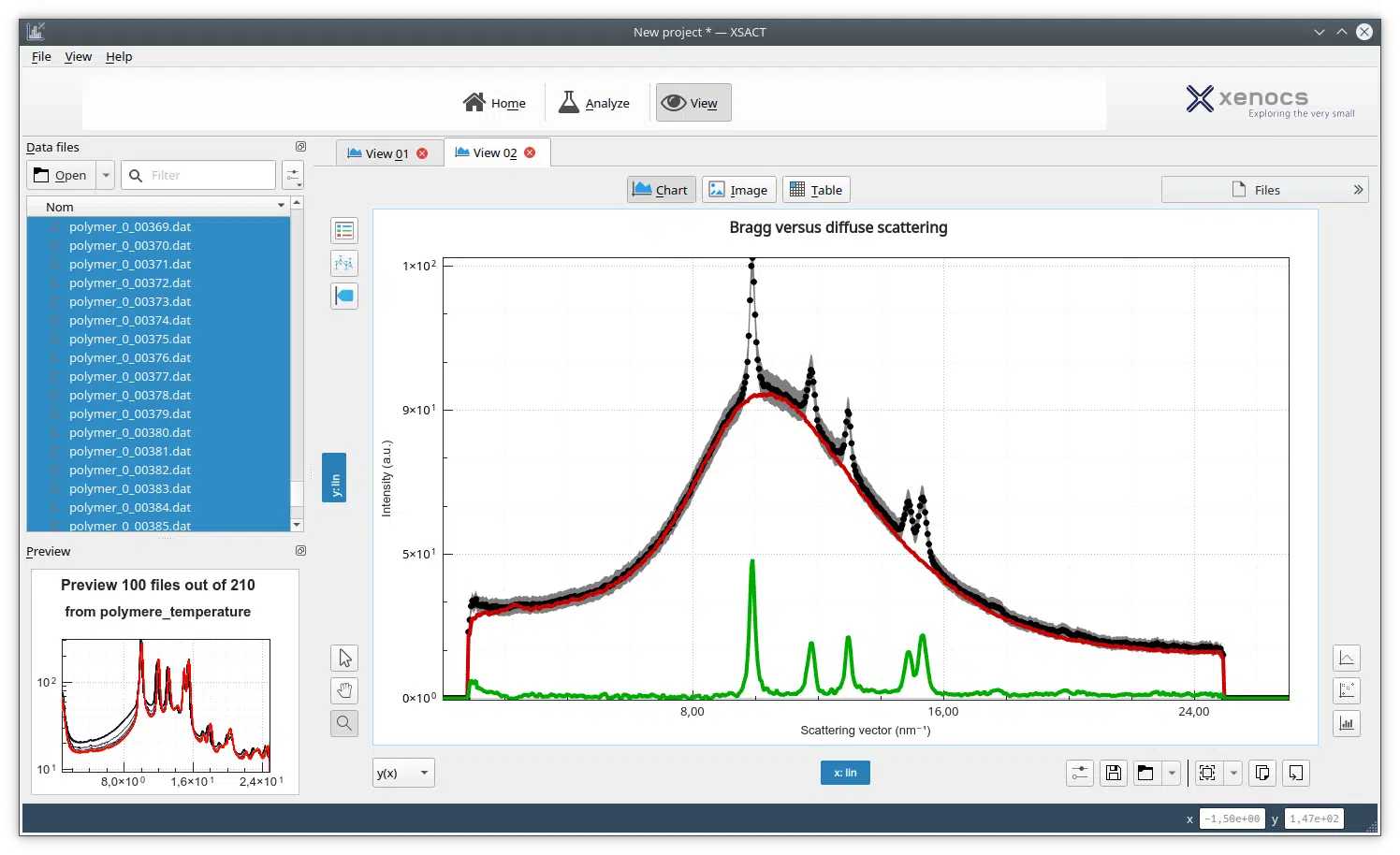
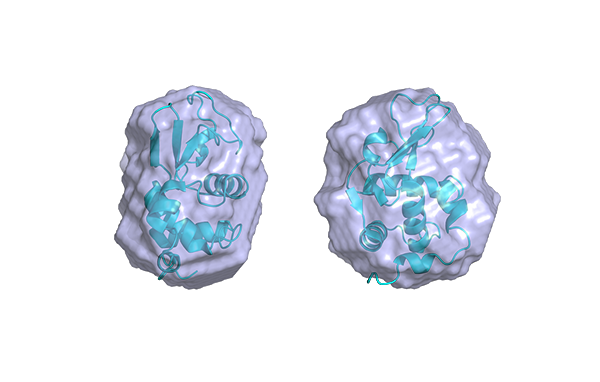
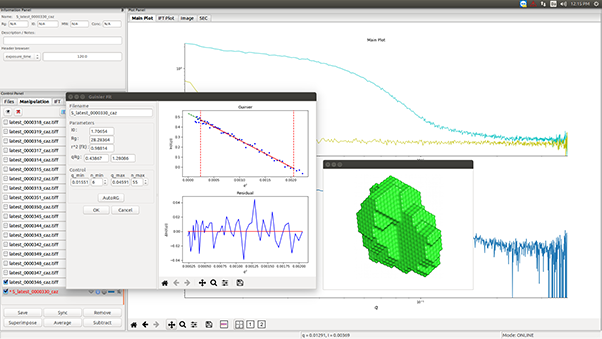
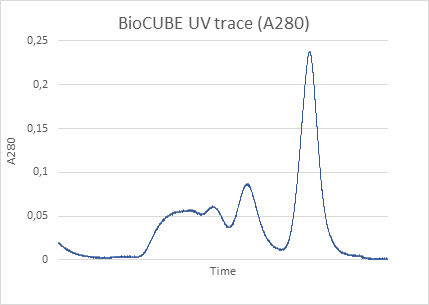
Small-Angle X-ray Scattering (SAXS) is a well-established technique that provides detailed insights into the size, shape, structure, stability, flexibility, oligomeric state, folding states, and interparticle interactions of samples in a near-native environment [9, 10]. Unlike traditional light scattering methods, which primarily measure particle size and polydispersity, SAXS offers specific structural insights into conformational states and flexibility, making it particularly valuable for studying complex proteins like antibodies. Additionally, SAXS’s high-throughput capabilities enable rapid data acquisition and analysis, making it an efficient tool for structural and biophysical characterization, as well as for validating models from crystallography, NMR, and computational predictions [11, 12].