The FEBS Journal, 2019, vol 0, 0,
DOI:10.1111/febs.15010
Abstract
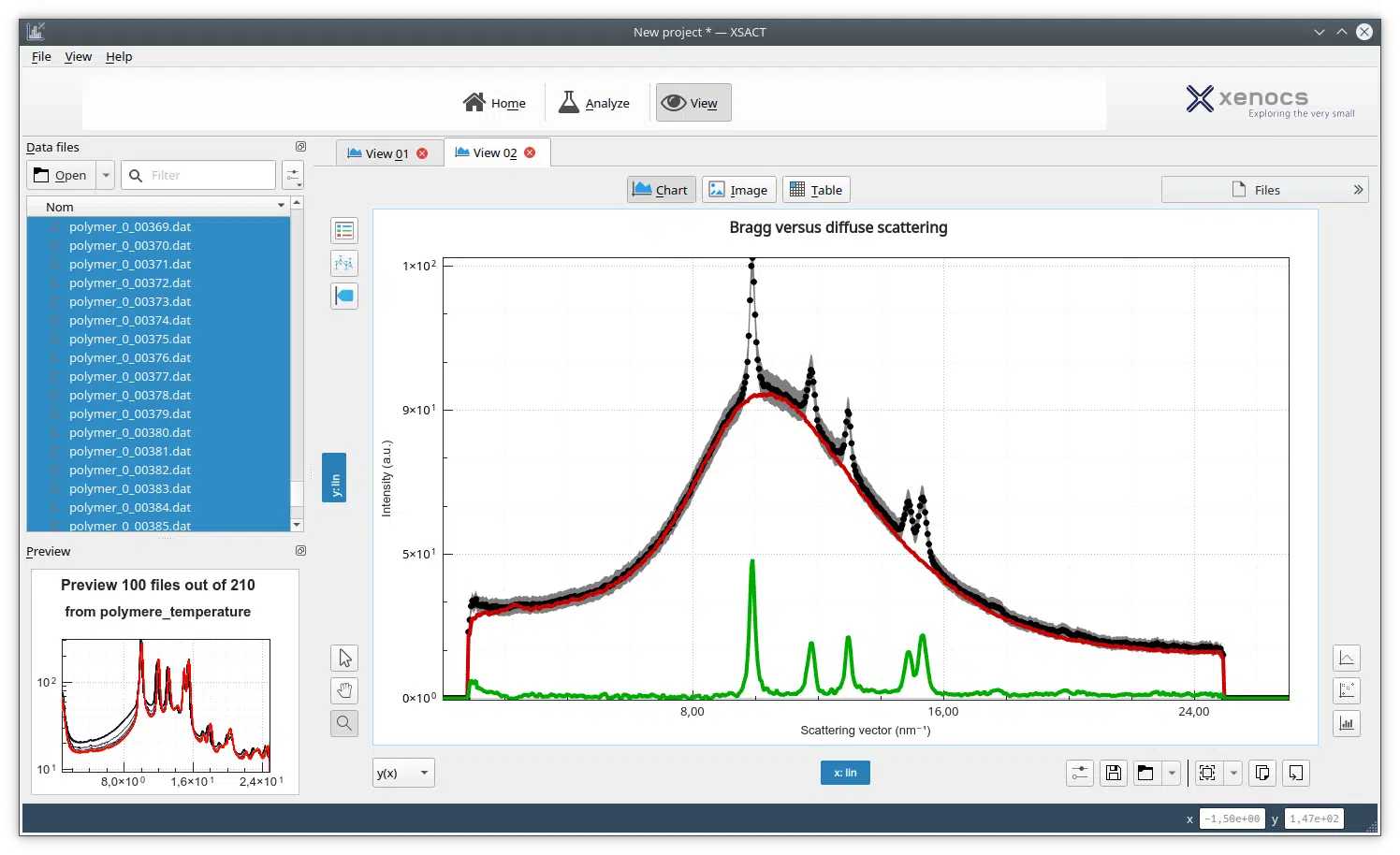
Human cystatin C (hCC), a member of the superfamily of papain-like cysteine protease inhibitors, is the most widespread cystatin in human body fluids. This small protein, in addition to its physiological function, is involved in various diseases, including cerebral amyloid angiopathy, cerebral hemorrhage, stroke, and dementia. Physiologically active hCC is a monomer. However, all structural studies based on crystallization led to the dimeric structure formed as a result of a three-dimensional exchange of the protein domains (3D domain swapping). The monomeric structure was obtained only for hCC variant V57N and for the protein stabilized by an additional disulfide bridge. With this study, we extend the number of models of monomeric hCC by an additional hCC variant with a single amino acid substitution in the flexible loop L1. The V57G variant was chosen for the X-ray and NMR structural analysis due to its exceptional conformational stability in solution. In this work, we show for the first time the structural and dynamics studies of human cystatin C variant in solution. We were also able to compare these data with the crystal structure of the hCC V57G and with other cystatins. The overall cystatin fold is retained in the solute form. Additionally, structural information concerning the N terminus was obtained during our studies and presented for the first time. Database Crystallographic structure: structural data are available in PDB databases under the accession number 6ROA. NMR structure: structural data are available in PDB and BMRB databases under the accession numbers 6RPV and 34399, respectively.