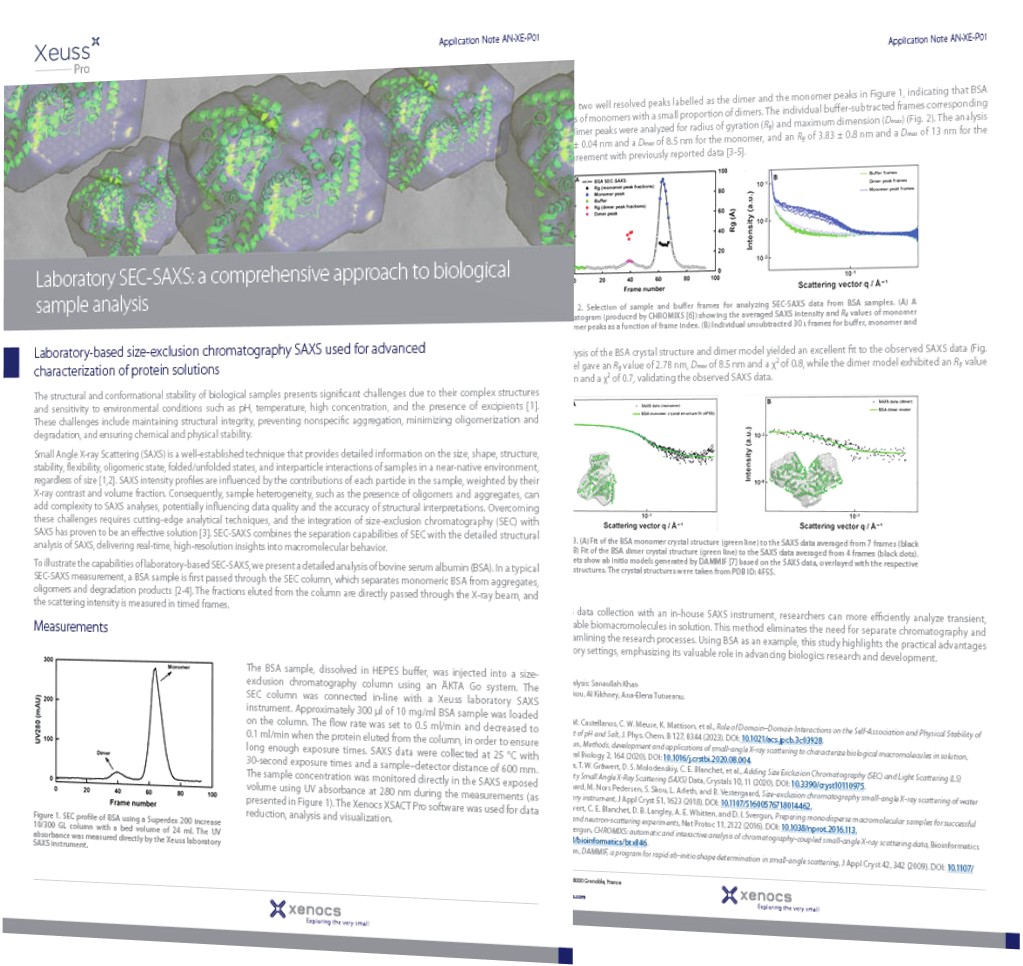
Laboratory-based size-exclusion chromatography SAXS used for advanced characterization of protein solutions
The structural and conformational stability of biological samples presents significant challenges due to their complex structures and sensitivity to environmental conditions such as pH, temperature, high concentration, and the presence of excipients [1]. These challenges include maintaining structural integrity, preventing nonspecific aggregation, minimizing oligomerization and degradation, and ensuring chemical and physical stability.
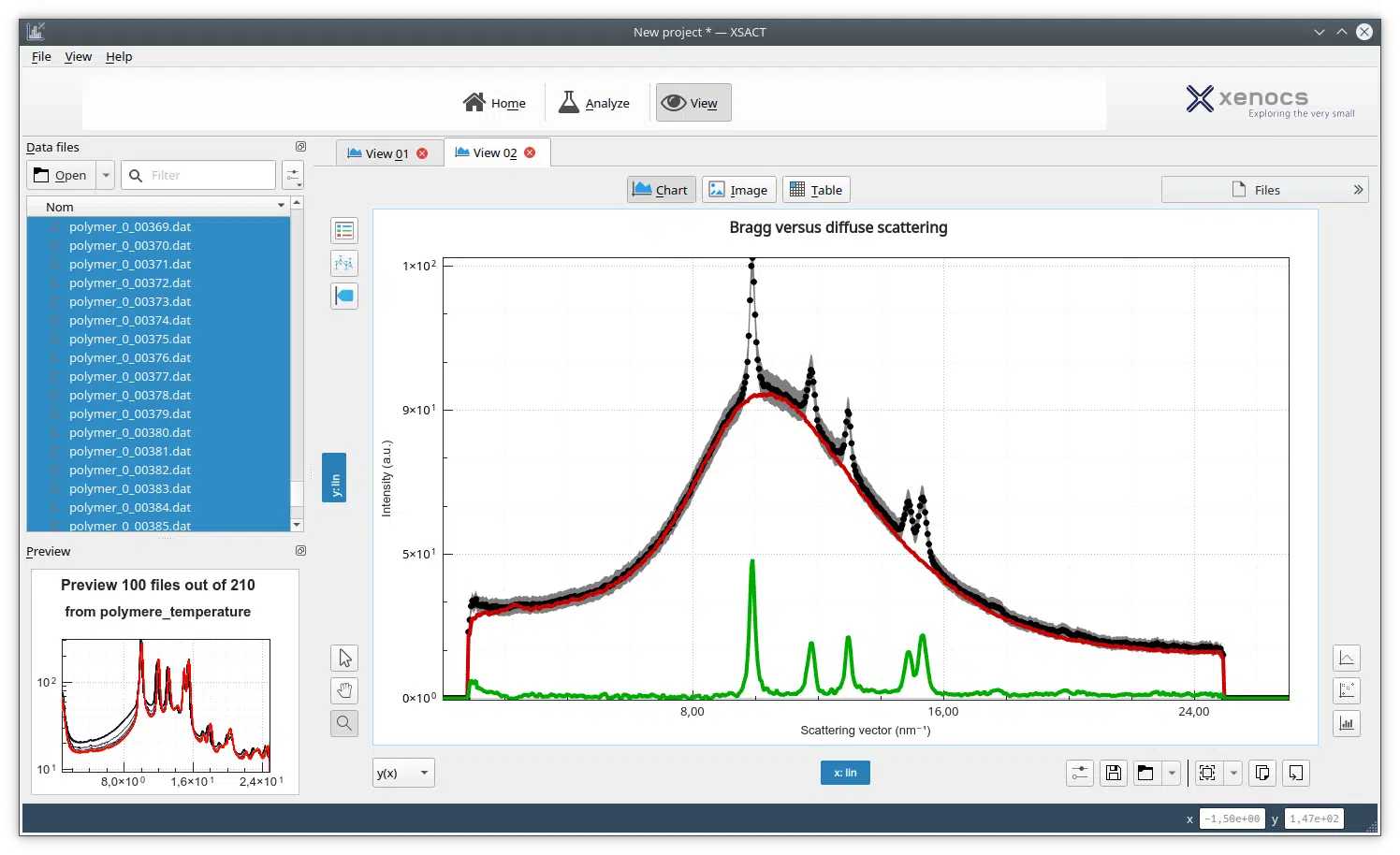
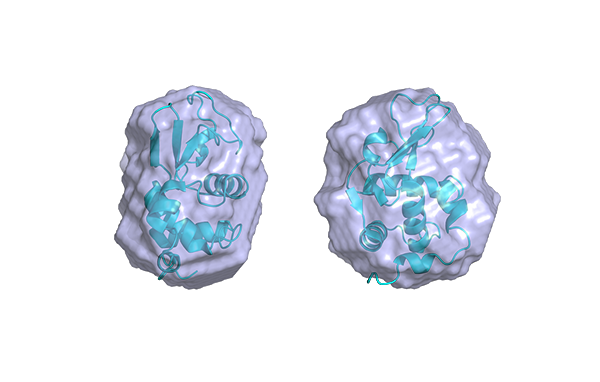
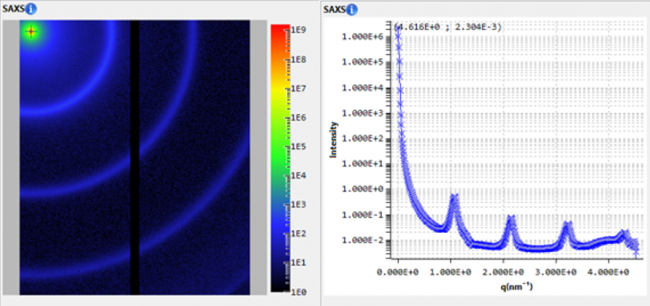
Small-Angle X-ray Scattering (SAXS) is a well-established technique that provides detailed information on the size, shape, structure, stability, flexibility, oligomeric state, folded/unfolded states, and interparticle interactions of samples in a near-native environment, regardless of size [1,2]. SAXS intensity profiles are influenced by the contributions of each particle in the sample, weighted by their X-ray contrast and volume fraction. Consequently, sample heterogeneity, such as the presence of oligomers and aggregates, can add complexity to SAXS analyses, potentially influencing data quality and the accuracy of structural interpretations. Overcoming these challenges requires cutting-edge analytical techniques, and the integration of size-exclusion chromatography (SEC) with SAXS has proven to be an effective solution [3]. SEC-SAXS combines the separation capabilities of SEC with the detailed structural analysis of SAXS, delivering real-time, high-resolution insights into macromolecular behavior.
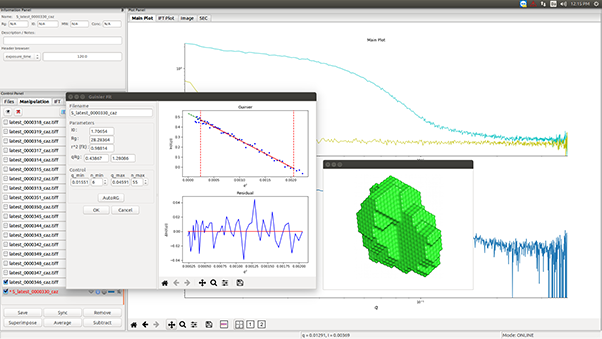
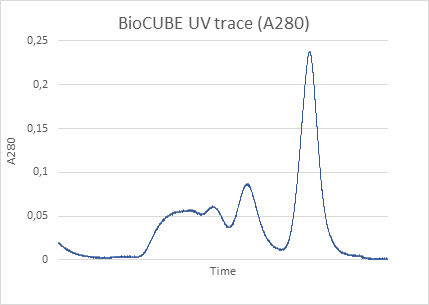
To illustrate the capabilities of laboratory-based SEC-SAXS, we present a detailed analysis of bovine serum albumin (BSA). In a typical SEC-SAXS measurement, a BSA sample is first passed through the SEC column, which separates monomeric BSA from aggregates, oligomers and degradation products [2-4]. The fractions eluted from the column are directly passed through the X-ray beam, and the scattering intensity is measured in timed frames.