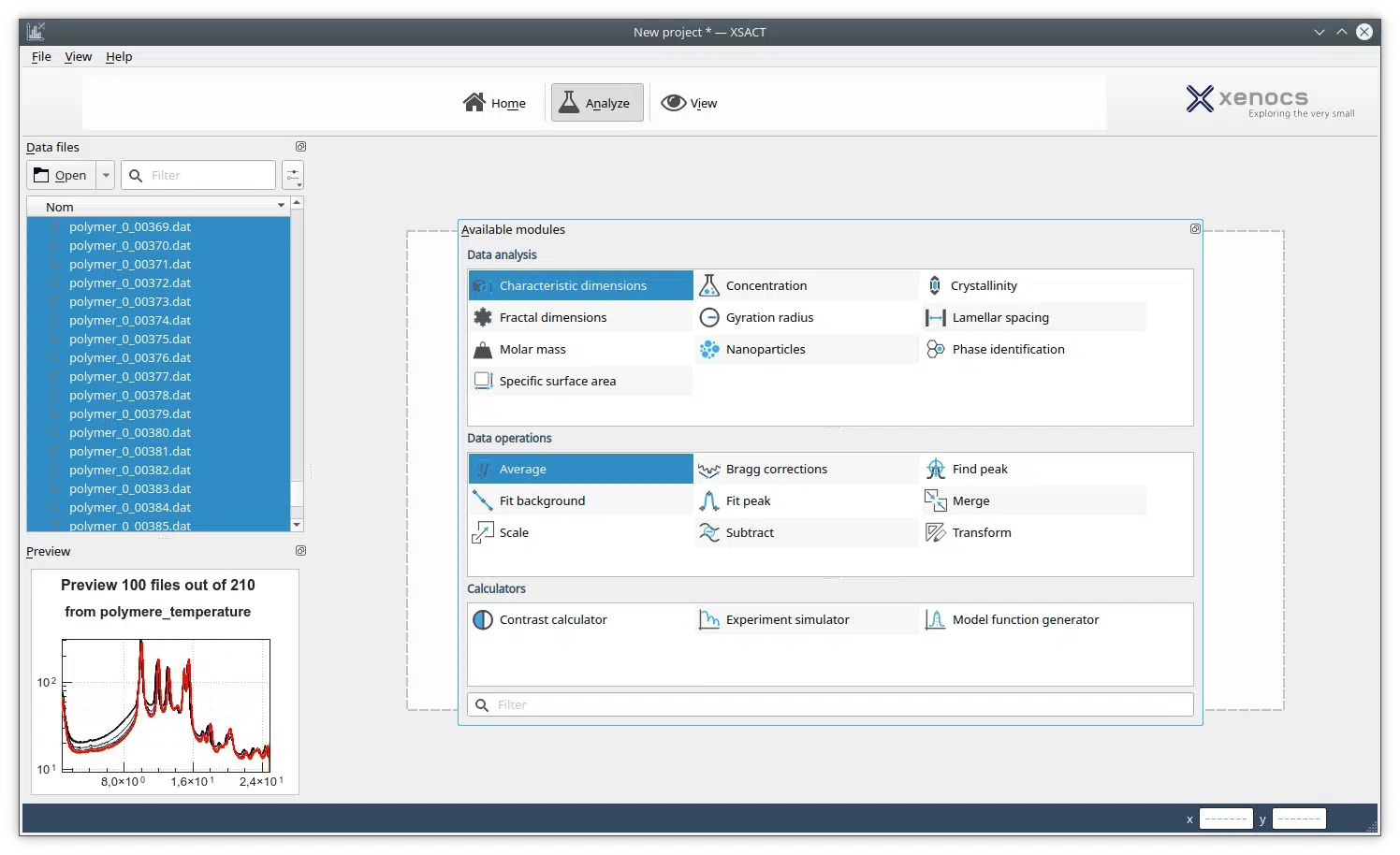
Particle or macromolecular shape
Particle or macromolecular shape, what is measured?
The pair distance distribution function (PDDF) is a histogram of distances within a single particle, for monodisperse, low concentration systems. The profile of the PDDF gives an indication of the shape of the particle (globular, prolate or oblate) as well as the dimensions of these particles.
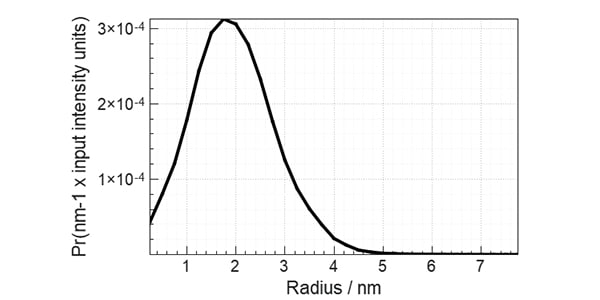
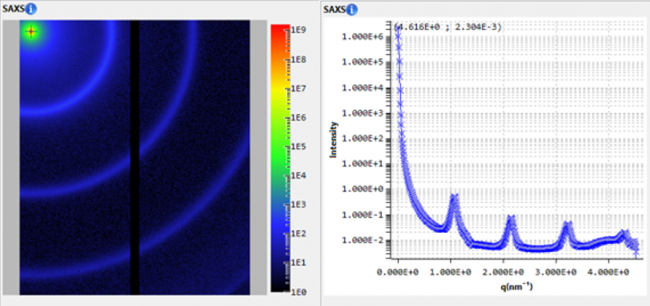
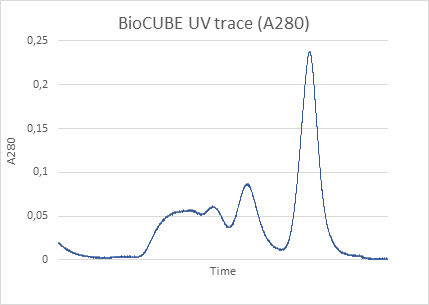
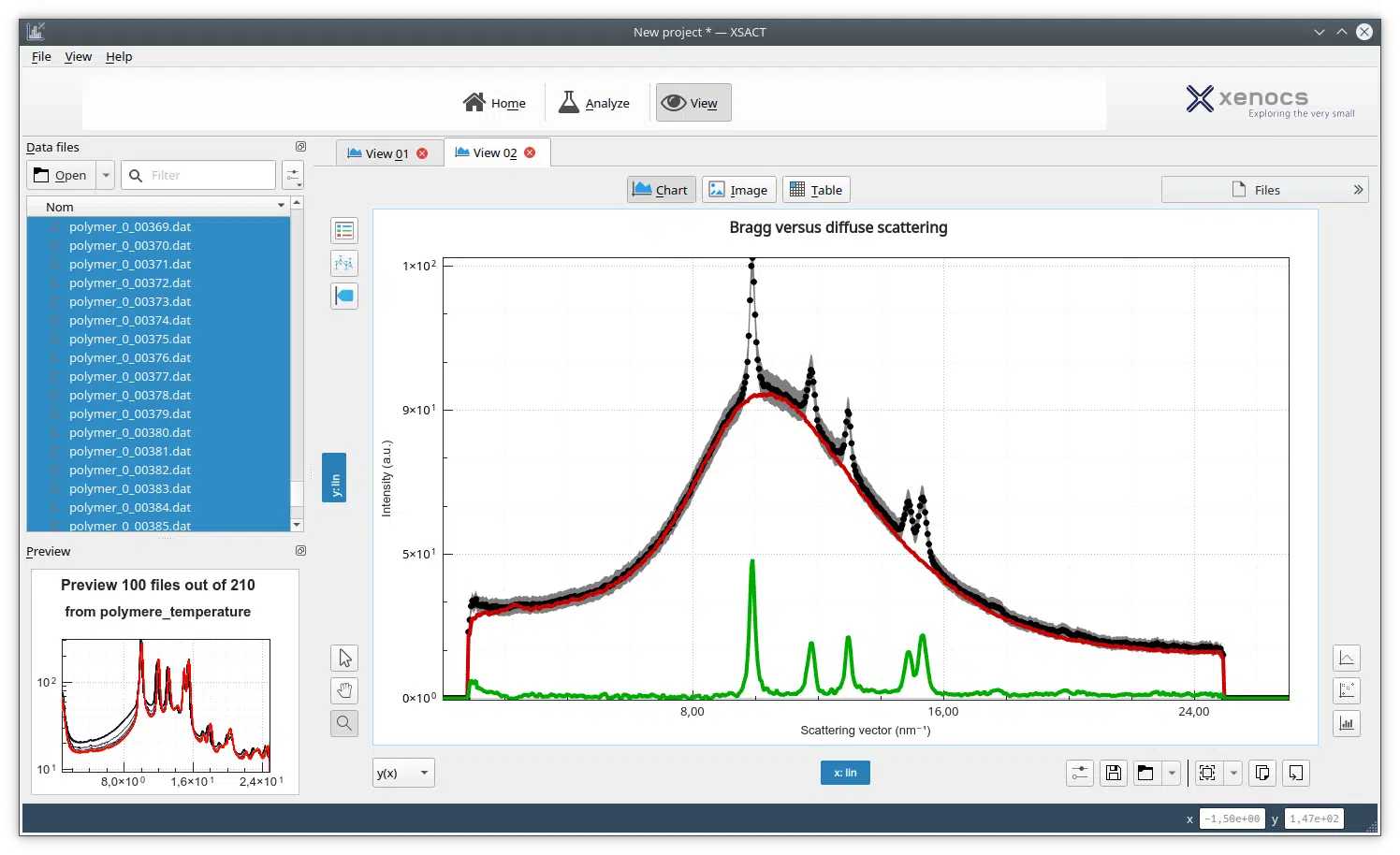
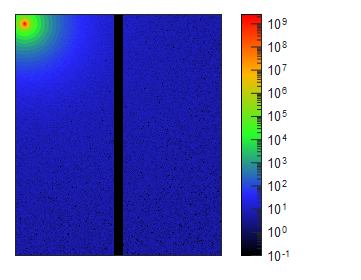
Figure 1. The pair distance distribution function obtained with low angle scattering data from lysozyme in aqueous solution, treated in XSACT. PDDF is obtained through a Bayesian approach, with no information loss. The user input of auxiliary values such as the maximum distance within the particle (Dmax) of the smoothness parameter are not required.
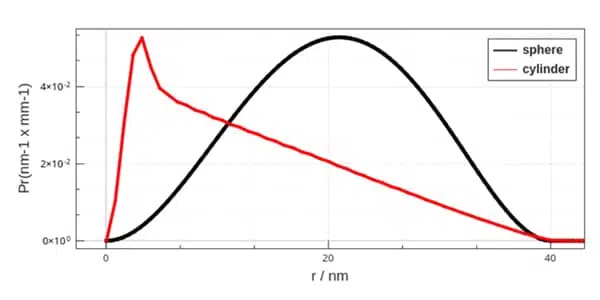
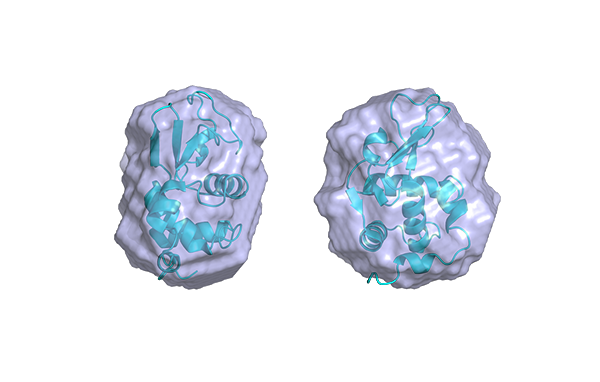
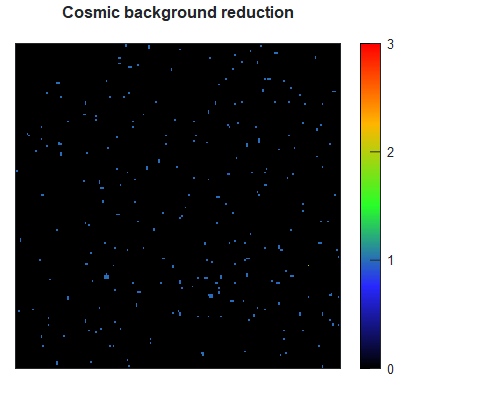
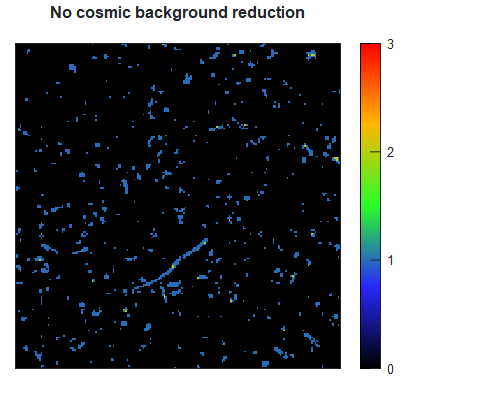
Figure 2. The profile of the PDDF hints to the shape of the particle. Comparison between the PDDF of spheres of 40 nm in diameter versus the one of a cylinder of 40 nm length and 4 nm diameter.
Samples
Typical samples for this measurement are:
-
Protein solutions
-
Dilute nanoparticles
-
Surfactants, micelles
-
Lipid based colloidal systems
Methods & standards
Standards & methods used for pair distance distribution function determination:
- Hansen, “Bayesian estimation of hyperparameters for indirect Fourier transformation in small‐angle scattering”, Journal of Applied Crystallography, vol. 33, pp 1415-1421, Dec. 2000
Example of customer publications making use of PDDF for sample analysis:
- Schillén, K. et al. “Block copolymers as bile salt sequestrants: intriguing structures formed in a mixture of an oppositely charged amphiphilic block copolymer and bile salt.” Phys. Chem. Chem. Phys. 21, 12518–12529 (2019).
XSACT analysis software implements the above methods.
Why use SAXS for particle or macromolecular shape determination?
Advantages of SAXS for particle or macromolecular shape determination:
-
Statistical relevance: the measured quantity is averaged throughout the illuminated volume
-
Non-destructive measurement technique
-
Measurement in native conditions (in solutions)
Products
All these measurements are possible directly in your lab.

Xeuss Pro
The Ultimate Solution for Nanoscale Characterization using SAXS/WAXS/GISAXS/USAXS/Imaging