Editorial
This special edition of ‘Exploring the very small’ is dedicated to Structural Biology.
You will find in it a selection of articles showing what is done today with Small Angle X-ray Scattering in biostructural research, together with some latest news on new product developments & innovations in SAXS instrumentation for biostructural research.
We hope you will enjoy ‘Exploring the very small’.
Xenocs Editorial Team
Today at the nanoscale
We are proud to highlight some of the recent publications from our customers using SAXS laboratory characterization for biostructural research. For a more complete list of publications related to this field please refer to our customer publications webpage using the bioscience filters.
Image credit : SAXS 3D envelope extracted from doi:10.1038/s41598-018-20835-x, figure 5.
S. Abehsera, S. Zaccai, B. Mittelman, L. Glazer, S. Weil, I. Khalaila, G. Davidov, R. Bitton, R. Zarivach, S. Li, F. Li, J. Xiang, R. Manor, E. D. Aflalo & A. Sagi; Scientific Reports, volume 8, Article number: 2430 (2018); doi:10.1038/s41598-018-20835-x
M.M. Castellanos, J. A. Snyder, M. Lee, S. Chakravarthy, N. J. Clark, A. McAuley, J.E.Curtis; Antibodies, 2017, 6(4), 25; doi:10.3390/antib6040025
Y. Sun, J. Zhang, H. Wang, T. Wang, H. Chen, B. Yu, C.L.P. Oliveira; Food hydrocolloids, vol 82, September 2018, Pages 19-28; doi:10.1016/j.foodhyd.2018.03.043
Any news which has raised your interest you want to share ? Please send it to us !
Characterization focus
Protein-protein interactions - from in vivo functionality to pharmaceutics and food science
Protein-protein interactions are relevant for various biological processes, such as transcription and signal transduction, but also for protein crystallization, stability and shelf-life of pharmaceutical formulations, as well as for processing and storage of protein-based food.
While repulsive interactions might stabilize a formulation, they will prevent protein crystallization which requires moderate attraction between proteins. A simple measure of weak pair-wise protein interactions is the osmotic second virial coefficient, A2, positive for repulsive interactions and negative in the case of attractions. It is traditionally determined using static light scattering (SLS) at low protein concentrations with a process that can quickly become tedious or require large sample volumes for even smaller parameter screenings.
SAXS eliminates many of the issues associated with obtaining A2 from SLS (Static Light Scattering). On the BioXolver, measuring A2 can be obtained with only 10 µL of sample per measurement with a very high degree of automation, even handling the sample mixing itself. The integrated UV-Vis spectrum is recorded to calculate the very important protein concentration used in the A2 measurements. This facilitates very large screening measurements that were previously unpractical or impossible.
As an example, the 2nd virial coefficient dependence on salt concentration for Lysozyme was examined using the Xenocs BioXolver.
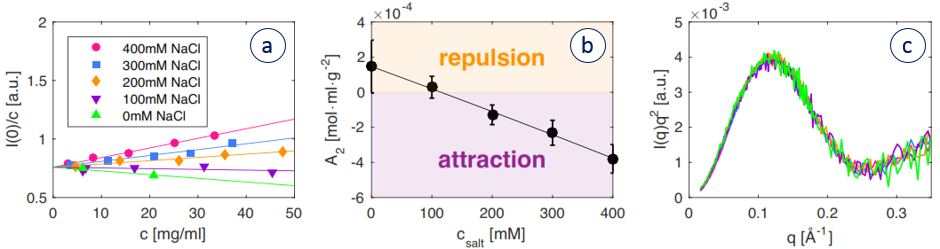
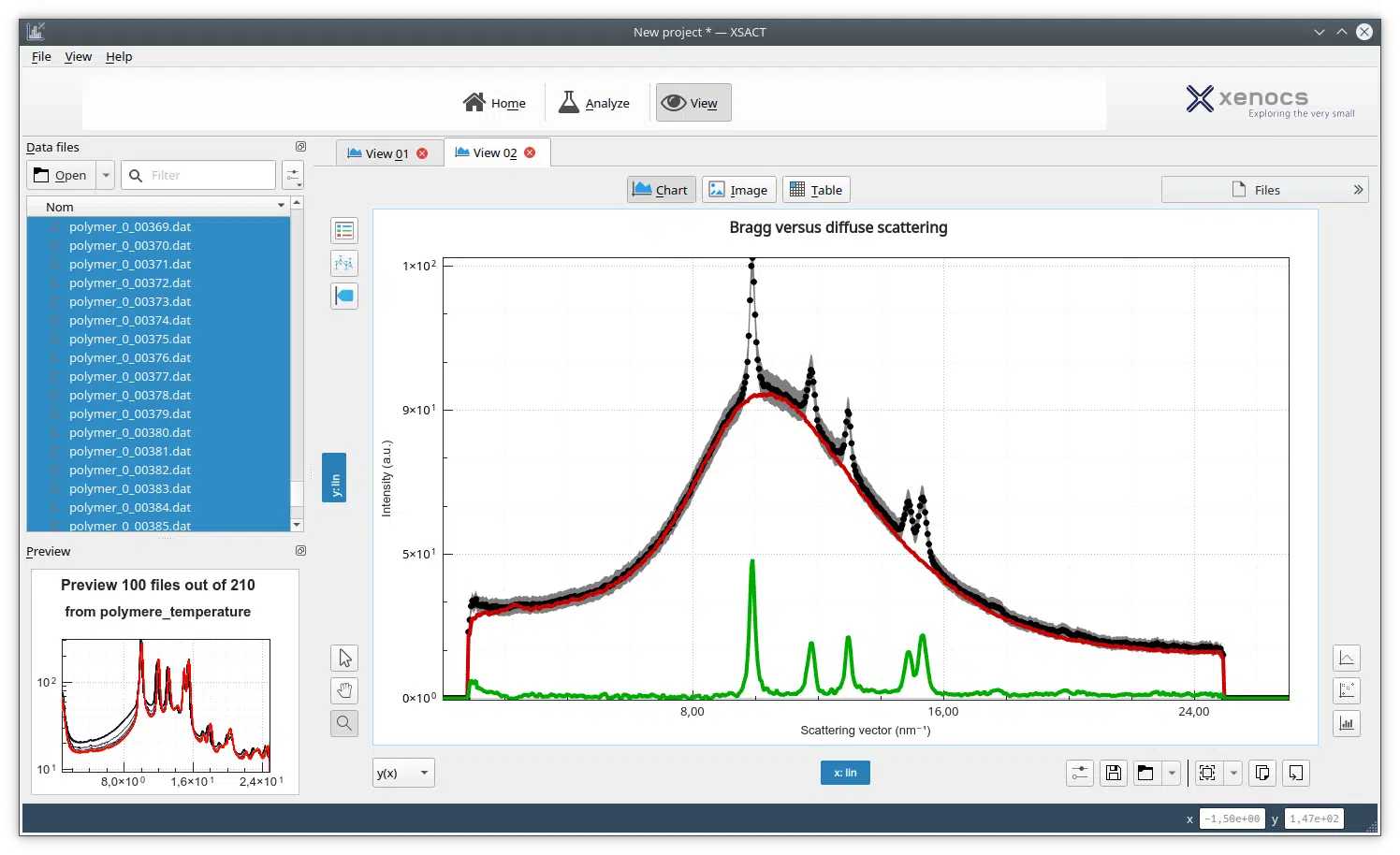
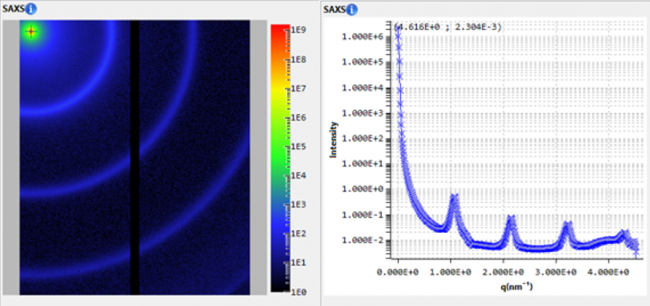
Figure 1. Forward scattering (a) and second virial coefficient (b) of lysozyme as a function of NaCl concentration. c: Kratky plot of 5 mg/ml lysozyme at varying salt concentration.
Figure 1 (a) shows the forward scattering points for different concentrations of salt and lysozyme concentration. From these data the second virial coefficient, Figure 1 (b), is obtained showing salt content above 100 mM leads to attraction in the sample. Figure 1 (c) displays the Kratky plot indicating a globular folded protein for the different salt concentrations. Had crystallization been the main goal, keeping the protein folded would have been important as well.
For protein crystallization mapping the value of A2 as a function of salt concentration allows researchers to dial a formulation in the so called “crystallization slot” loosely defined between -1 and -8 x 10-4 mol·mL/g2 where the protein interaction are just enough to form diffracting crystals.
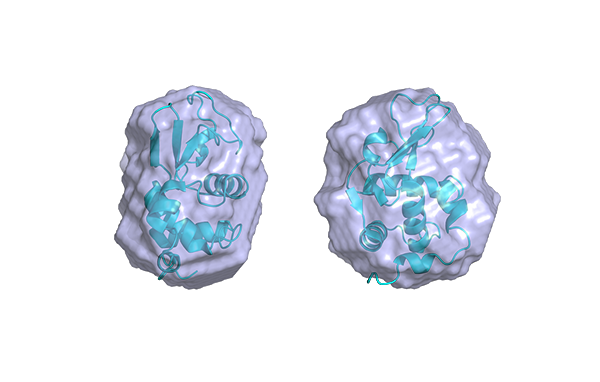
For more information and another example of protein interactions showing the unfolding of Bovine serum albumin as a function of buffer pH, shown in figure 2, please see our application note on the website.
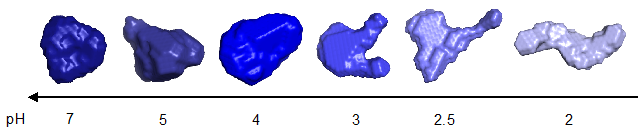
Figure 2. Bovine Serum Albumin shape change as a function of pH.
They are exploring the very small
Cancer treatment: studying the function & regulation of PARPs proteins with SAXS
Interview with Associate Professor John Pascal, Department of Biochemistry and Molecular Medicine, Faculty of Medicine, Université de Montréal, Canada.

Could you please briefly introduce your research in structural biology?
J. Pascal : A broad research interest of my group is the cellular response to DNA damage. We study the proteins that detect and signal the presence of DNA damage, as well as the enzymes that repair the DNA damage. A rapid and efficient cellular response to DNA damage is critical to the integrity of the genome. Our research is aimed toward understanding how these proteins/enzymes work at the molecular level. We also have a strong interest in the family of proteins known as poly(ADP-ribose) polymerases, or PARPs. PARPs are involved in regulating virtually every aspect of human cell biology, and several PARP enzymes are targets for cancer treatment. Overall, we combine biochemical analysis, cell-based experiments, and structural biology techniques to advance our basic understanding of protein function, and we also work to leverage this structural information toward the development of new types of inhibitors that could potentially serve as therapeutics.
What are you using SAXS in your research for ?
J. Pascal : We have used SAXS to address the overall shape and conformation of macromolecules and macromolecular complexes in solution. We are enthusiastic about SAXS! It has been particularly useful for the highly modular and flexible proteins that we have investigated. Whereas x-ray crystallography has often provided us with views of the more stable portions of a protein, SAXS analysis has often been able to capture information on full-length proteins and provide insights into the overall flexibility and relative positioning of the component parts/domains of the protein. We have also used SAXS to measure changes in protein conformation in response to interaction with small molecule inhibitors and protein binding partners.
What benefits does an in-house SAXS instrument have, compared with using a synchrotron facility ?
J. Pascal : An in-house system provides the ability to respond in real time to the results, preparing new samples on the fly in response to what you are observing. This ability is often key to finding the right conditions for a particular sample.
There is also the question of how to ship the samples to the synchrotron, whether to send with a freeze then thaw cycle, or to just send as a fluid on ice. Not all samples behave well under these shipment conditions. The in-house source allows us to collect data when the sample is most “fresh” and ready-to-go. We have also used an in-house system to test for the best storage conditions so that we could then ship to synchrotrons more confidently.
Synchrotrons will always be an important and instrumental part of our work, but an in-house system helps us to properly prepare for optimal use of synchrotron facilities. We have had some proteins that suffer dramatically from synchrotron radiation damage, and an in-house source of lower intensity radiation has helped in these cases.
Why did you choose to buy the BioXolver from SAXSLAB/Xenocs ?
J. Pascal : We liked that the capabilities of the system are quite adaptable to the needs of the experiment. We felt that we would never be limited in the types of experiments that we could perform.
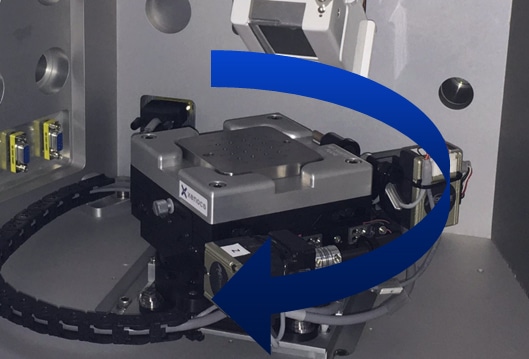
The machine is able to get through a substantial number of samples in a day’s time. The robotic sample loading and the visual feedback on sample positioning in the beamline are particularly nice. Precious and difficult to purify samples can be loaded in fairly small volumes that we can confidently keep in the beamline during the course of the experiment.
Product highlight
Accelerate your Biostructural research with BioXolver
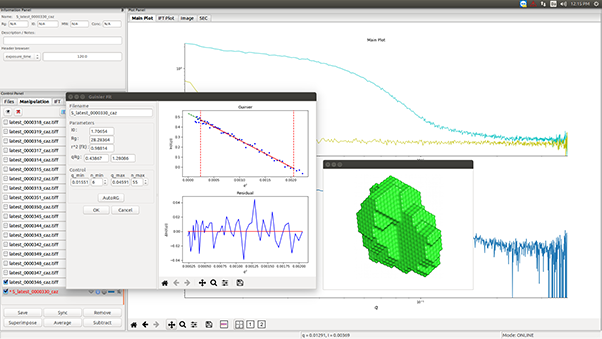
The BioXolver is designed for biologists who require fast and easy access to high quality information on the shape and dynamics of macromolecules in solution. It uses Small Angle X-ray Scattering (SAXS) to provide information on structure and conformation of biological macromolecules and complexes over a wide range of dimensions (down to 1 nm) while the sample is maintained in its natural environment. Lipid membranes, protein-protein interactions, globular, flexible and intrinsically disordered proteins and enzymes, fibrillation, microgels are just a few examples of fields of study where SAXS characterization can be uniquely beneficial. Valuable sample parameters such as radius of gyration, molecular weight, maximum intra-particle distance, degree of folding and the low-resolution 3D protein shape are easily accessible with little sample preparation and high throughput.

Optimized for maximum X-ray intensity and lowest background noise, the BioXolver delivers excellent sensitivity and data quality even for highly diluted or low molecular weight proteins and reduces or even eliminates the need for synchrotron beamtime. Data from the BioXolver can also be used in synergy with other techniques for validation of high resolution structures obtained by intrusive methods such as cryo-EM, protein crystallography, or for hybrid structure modelling of complexes or assemblies combining data from several different techniques [1].
The BioXolver offers maximum throughput. Up to 192 samples can be loaded and measured automatically using an in-line pipetting robot and the automated software suite. The conjunction of the in-line pipetting robot and machine vision ensures a total sample consumption as low as 5µL per sample – a feature which could significantly lower the cost of the measurements. This unique combination of low cost/volume of sample and high level of automation makes it possible to access, on a daily basis, information in in-vivo conditions. Such a contribution could reduce the R&D cycle for biostructural investigations, including protein formulation [2] and crystallization studies.
[1] Vestergaard B. & Al.; Arch Biochem Biophys. 2016 Jul 15;602:69-79
[2] X. Tian & Al.; J. Pharm. Sci. 103 (2014) p1701-1710
Xenocs news
First stone laid for Xenocs new Head Office
Last April Xenocs launched the construction of its new head office in Grenoble. The new facility will allow Xenocs to concentrate all its Grenoble-based activities in one site and support further growth for years to come. It will host Xenocs production and R&D activities and provide the team at Xenocs with a modern and stimulating work environment.

“The new building was designed with our customers and our team in mind. It is a major step in the development of the company, enabling us to sustain the growth Xenocs has experienced over the last years. Innovation and quality are at the heart of our company and the new facility will be the perfect framework for providing just that to our customers” Xenocs CEO Peter Høghøj states.
Located within walking distance from several large international research facilities such as the European Synchrotron Radiation Facility (ESRF), Xenocs will benefit from the dynamic area. The new site will be a center of excellence for the training of Xenocs customers and will welcome potential customers in its state-of-the-art laboratory.
“Xenocs new head office is part of our strategy to come closer to our customers. Not only it will be easily accessible from airports & train stations, but also it will be located next to world-class research centers visited by many of our customers. This will be the opportunity for our customers to combine a visit to the Synchrotron with the ability to come & discuss with our SAXS experts and potentially make measurements in our lab”, Xenocs Managing Director Frédéric Bossan says.
Opening is planned for September 2019.