Researchers from Université Laval successfully used the SEC-SAXS laboratory setup at the Structural Biology Platform of Université de Montréal to elucidate the structure of PieE. This enzyme is involved in the biosynthesis of the antibiotic piericidin. It facilitates the hydroxylation of a substrate with the help of NADPH, O2, PAD and C4a-hydroperosxyflavin intermediate formation. Interestingly, this enzyme possesses a control system to prevent the oxidation of NADPH and the reduction of FAD in the absence of the substrate. The current publication M.S. Manenda et al.1 sheds a light on the molecular basis of this control system with the help of Size-Exclusion Chromatography coupled with Multi-Angle Light Scattering (SEC-MALS and Small-Angle X-ray Scattering (SEC-SAXS) in combination with protein crystallography and an activity assays.
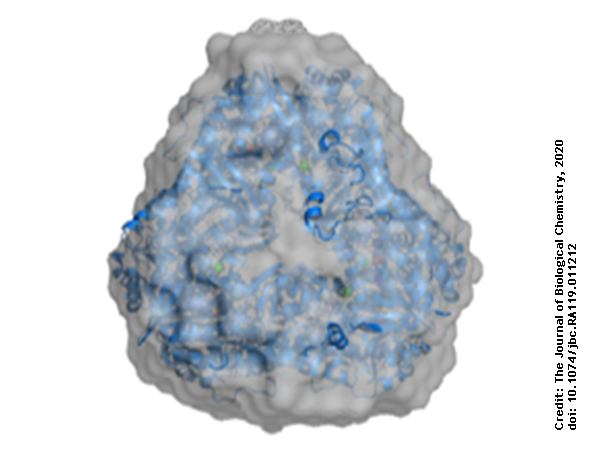
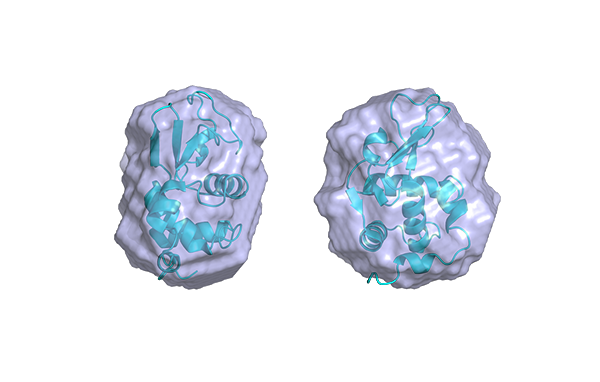
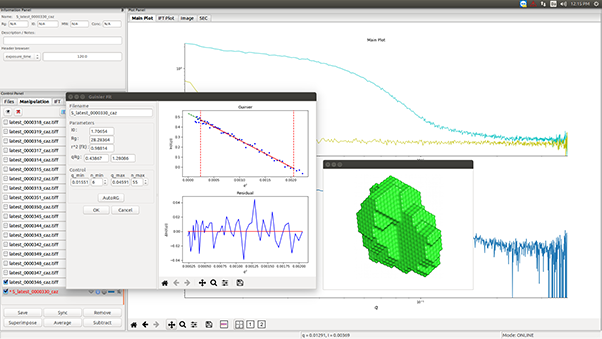
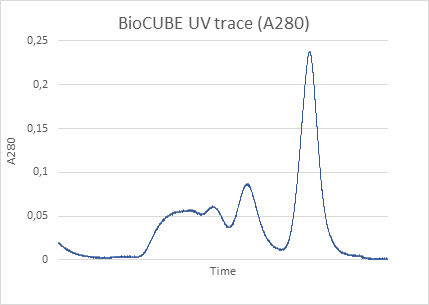
Using protein crystallography, PieE was shown to form a hexameric structure in three different crystal forms. Subsequently, SEC-MALS and SEC-SAXS were used to determine the absolute molecular mass and to confirm the presence of the oligomeric state in solution. Moreover, SAXS provided additional structural information: the radius of gyration and the maximum dimensions of the enzyme in solution were determined with this technique. Using ab initio shape reconstruction, the researchers composed a dummy atom model fitting (picture) which showed a good agreement in both the shape and the volume with the crystal structure.
This combination of techniques provided a robust determination of the structure of PieE in solution and in the crystal form, which forms a solid basis for the subsequent understanding of the catalytic mechanism. A combination of crystallography of different states and activity assays of the wild type and mutants of PieE was then used to unveil essential steps and conformations in catalytic cycle that are of importance for the whole class of Group A flavin-dependent monooxygenases, revealing an additional sliding conformation existing in between the classical IN and OUT conformations.